* Migrate to bzlmod
* Update Python version to PY3, as indicated by the actual source file.
* Migrate more libraries & first draft of direct pywheel rule usage in Bazel
* Integrate with nanobind and libpfm
* Make Python toolchain a dev dependency
* Undo py_wheel usage until later
* Added support for bzlmod for C++ parts of google_benchmark.
* Make //tools:all buildable with --enable_bzlmod
---------
Co-authored-by: Andy Christiansen <achristiansen@google.com>
Previously compare.py ignored the --benchmarks_filter
argument when loading JSON. This defeated any workflow when
a single run of the benchmark was run, followed by multiple
"subset reports" run against it with the 'benchmarks'
command.
Concretely this came up with the simple case:
compare.py benchmarks a.json b.json --benchmarks_filter=BM_Example
This has no practical impact on the 'filters' and
'benchmarkfiltered' comand, which do their thing at a later
stage.
Fixes#1484
Co-authored-by: dominic <510002+dmah42@users.noreply.github.com>
* Build libpfm as a dependency to allow collection of perf counters
This commit builds libpfm using rules_foreign_cc and lets the default
build of the benchmark library support perf counter collection without
needing additional work from users.
Tested with a custom target:
```
bazel run \
--override_repository=com_github_google_benchmark=/home/raghu/benchmark \
-c opt :test-bench -- "--benchmark_perf_counters=INSTRUCTIONS,CYCLES"
Using profile: local
<snip>
----------------------------------------------------------------------
Benchmark Time CPU Iterations UserCounters...
----------------------------------------------------------------------
BM_Test 0.279 ns 0.279 ns 1000000000 CYCLES=1.00888 INSTRUCTIONS=2
```
Signed-off-by: Raghu Raja <raghu@enfabrica.net>
* Adding myself to the CONTRIBUTORS file per CLA guidance
Enfabrica has already signed a corporate CLA.
Signed-off-by: Raghu Raja <raghu@enfabrica.net>
Signed-off-by: Raghu Raja <raghu@enfabrica.net>
Some linux distributions no longer provide `python` binary and require
usage of `python3` instead. This changes the scripts here and uses
cmake `find_package(Python3` when running python.
Co-authored-by: Dominic Hamon <dominichamon@users.noreply.github.com>
The pandas.Timedelta class truncates to integral nanoseconds, which throws
away sub-nanosecond precision present in benchmark JSON. Switch to
floating point multiplication, which preserves it.
Fixes#1482
Tentatively fixes#1477.
* Add possibility to ask for libbenchmark version number (#1004)
Add a header which holds the current major, minor, and
patch number of the library. The header is auto generated
by CMake.
* Do not generate unused functions (#1004)
* Add support for version number in bazel (#1004)
* Fix clang format #1004
* Fix more clang format problems (#1004)
* Use git version feature of cmake to determine current lib version
* Rename version_config header to version
* Bake git version into bazel build
* Use same input config header as in cmake for version.h
* Adapt the releasing.md to include versioning in bazel
Despite the wide variety of the features we provide,
some people still have the audacity to complain and demand more.
Concretely, i *very* often would like to see the overall result
of the benchmark. Is the 'new' better or worse, overall,
over all the non-aggregate time/cpu measurements.
This comes up for me most often when i want to quickly see
what effect some LLVM optimization change has on the benchmark.
The idea is straight-forward, just produce four lists:
wall times for LHS benchmark, CPU times for LHS benchmark,
wall times for RHS benchmark, CPU times for RHS benchmark;
then compute geomean for each one of those four lists,
and compute the two percentage change between
* geomean wall time for LHS benchmark and geomean wall time for RHS benchmark
* geomean CPU time for LHS benchmark and geomean CPU time for RHS benchmark
and voila!
It is complicated by the fact that it needs to graciously handle
different time units, so pandas.Timedelta dependency is introduced.
That is the only library that does not barf upon floating times,
i have tried numpy.timedelta64 (only takes integers)
and python's datetime.timedelta (does not take nanosecons),
and they won't do.
Fixes https://github.com/google/benchmark/issues/1147
* Statistics: add support for percentage unit in addition to time
I think, `stddev` statistic is useful, but confusing.
What does it mean if `stddev` of `1ms` is reported?
Is that good or bad? If the `median` is `1s`,
then that means that the measurements are pretty noise-less.
And what about `stddev` of `100ms` is reported?
If the `median` is `1s` - awful, if the `median` is `10s` - good.
And hurray, there is just the statistic that we need:
https://en.wikipedia.org/wiki/Coefficient_of_variation
But, naturally, that produces a value in percents,
but the statistics are currently hardcoded to produce time.
So this refactors thinkgs a bit, and allows a percentage unit for statistics.
I'm not sure whether or not `benchmark` would be okay
with adding this `RSD` statistic by default,
but regales, that is a separate patch.
Refs. https://github.com/google/benchmark/issues/1146
* Address review notes
The test used to work with scipy 1.6, but does not work with 1.7.1
I believe this is because of https://github.com/scipy/scipy/pull/4933
Since there doesn't appear that there is anything wrong with how
we call the `mannwhitneyu()`, i guess we should just expect the new values.
Notably, the tests now fail with earlier scipy versions.
Currently, the tooling just keeps the whatever benchmark order
that was present, and this is fine nowadays, but once the benchmarks
will be optionally run interleaved, that will be rather suboptimal.
So, now that i have introduced family index and per-family instance index,
we can define an order for the benchmarks, and sort them accordingly.
There is a caveat with aggregates, we assume that they are in-order,
and hopefully we won't mess that order up..
NOTE: This is a fresh-start of #738 pull-request which I messed up by re-editing the commiter email which I forgot to modify before pushing. Sorry for the inconvenience.
This PR brings proposed solution for functionality described in #737Fixes#737.
* Create pylint.yml
* improve file matching
* fix some pylint issues
* run on PR and push (force on master only)
* more pylint fixes
* suppress noisy exit code and filter to fatals
* add conan as a dep so the module is importable
* fix lint error on unreachable branch
* add requirements.txt for python tools
* adds documentation for requirements.txt
Adds installation instructions for python dependencies using pip and requirements.txt
* Refactor U-Test calculation into separate function.
And implement 'print_utest' functionality in terms of it.
* Change 'optimal_repetitions' to 'more_optimal_repetitions'.
My knowledge of python is not great, so this is kinda horrible.
Two things:
1. If there were repetitions, for the RHS (i.e. the new value) we were always using the first repetition,
which naturally results in incorrect change reports for the second and following repetitions.
And what is even worse, that completely broke U test. :(
2. A better support for different repetition count for U test was missing.
It's important if we are to be able to report 'iteration as repetition',
since it is rather likely that the iteration count will mismatch.
Now, the rough idea on how this is implemented now. I think this is the right solution.
1. Get all benchmark names (in order) from the lhs benchmark.
2. While preserving the order, keep the unique names
3. Get all benchmark names (in order) from the rhs benchmark.
4. While preserving the order, keep the unique names
5. Intersect `2.` and `4.`, get the list of unique benchmark names that exist on both sides.
6. Now, we want to group (partition) all the benchmarks with the same name.
```
BM_FOO:
[lhs]: BM_FOO/repetition0 BM_FOO/repetition1
[rhs]: BM_FOO/repetition0 BM_FOO/repetition1 BM_FOO/repetition2
...
```
We also drop mismatches in `time_unit` here.
_(whose bright idea was it to store arbitrarily scaled timers in json **?!** )_
7. Iterate for each partition
7.1. Conditionally, diff the overlapping repetitions (the count of repetitions may be different.)
7.2. Conditionally, do the U test:
7.2.1. Get **all** the values of `"real_time"` field from the lhs benchmark
7.2.2. Get **all** the values of `"cpu_time"` field from the lhs benchmark
7.2.3. Get **all** the values of `"real_time"` field from the rhs benchmark
7.2.4. Get **all** the values of `"cpu_time"` field from the rhs benchmark
NOTE: the repetition count may be different, but we want *all* the values!
7.2.5. Do the rest of the u test stuff
7.2.6. Print u test
8. ???
9. **PROFIT**!
Fixes#677
As previously discussed, let's flip the switch ^^.
This exposes the problem that it will now be run
for everyone, even if one did not read the help
about the recommended repetition count.
This is not good. So i think we can do the smart thing:
```
$ ./compare.py benchmarks gbench/Inputs/test3_run{0,1}.json
Comparing gbench/Inputs/test3_run0.json to gbench/Inputs/test3_run1.json
Benchmark Time CPU Time Old Time New CPU Old CPU New
--------------------------------------------------------------------------------------------------------
BM_One -0.1000 +0.1000 10 9 100 110
BM_Two +0.1111 -0.0111 9 10 90 89
BM_Two +0.2500 +0.1125 8 10 80 89
BM_Two_pvalue 0.2207 0.6831 U Test, Repetitions: 2. WARNING: Results unreliable! 9+ repetitions recommended.
BM_Two_stat +0.0000 +0.0000 8 8 80 80
```
(old screenshot)
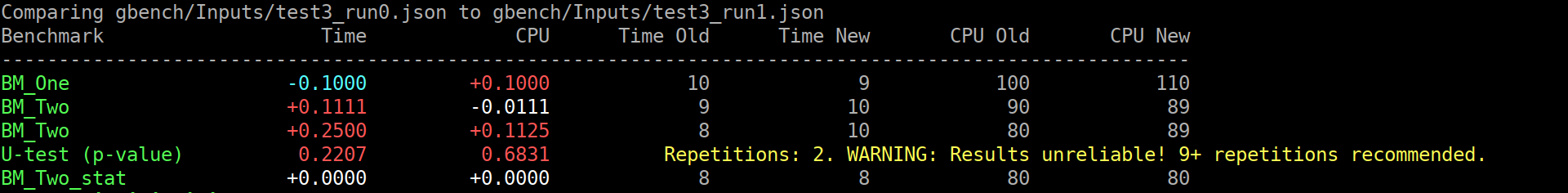
Or, in the good case (noise omitted):
```
s$ ./compare.py benchmarks /tmp/run{0,1}.json
Comparing /tmp/run0.json to /tmp/run1.json
Benchmark Time CPU Time Old Time New CPU Old CPU New
---------------------------------------------------------------------------------------------------------------------------------
<99 more rows like this>
./_T012014.RW2/threads:8/real_time +0.0160 +0.0596 46 47 10 10
./_T012014.RW2/threads:8/real_time_pvalue 0.0000 0.0000 U Test, Repetitions: 100
./_T012014.RW2/threads:8/real_time_mean +0.0094 +0.0609 46 47 10 10
./_T012014.RW2/threads:8/real_time_median +0.0104 +0.0613 46 46 10 10
./_T012014.RW2/threads:8/real_time_stddev -0.1160 -0.1807 1 1 0 0
```
(old screenshot)

The first problem you have to solve yourself. The second one can be aided.
The benchmark library can compute some statistics over the repetitions,
which helps with grasping the results somewhat.
But that is only for the one set of results. It does not really help to compare
the two benchmark results, which is the interesting bit. Thankfully, there are
these bundled `tools/compare.py` and `tools/compare_bench.py` scripts.
They can provide a diff between two benchmarking results. Yay!
Except not really, it's just a diff, while it is very informative and better than
nothing, it does not really help answer The Question - am i just looking at the noise?
It's like not having these per-benchmark statistics...
Roughly, we can formulate the question as:
> Are these two benchmarks the same?
> Did my change actually change anything, or is the difference below the noise level?
Well, this really sounds like a [null hypothesis](https://en.wikipedia.org/wiki/Null_hypothesis), does it not?
So maybe we can use statistics here, and solve all our problems?
lol, no, it won't solve all the problems. But maybe it will act as a tool,
to better understand the output, just like the usual statistics on the repetitions...
I'm making an assumption here that most of the people care about the change
of average value, not the standard deviation. Thus i believe we can use T-Test,
be it either [Student's t-test](https://en.wikipedia.org/wiki/Student%27s_t-test), or [Welch's t-test](https://en.wikipedia.org/wiki/Welch%27s_t-test).
**EDIT**: however, after @dominichamon review, it was decided that it is better
to use more robust [Mann–Whitney U test](https://en.wikipedia.org/wiki/Mann–Whitney_U_test)
I'm using [scipy.stats.mannwhitneyu](https://docs.scipy.org/doc/scipy/reference/generated/scipy.stats.mannwhitneyu.html#scipy.stats.mannwhitneyu).
There are two new user-facing knobs:
```
$ ./compare.py --help
usage: compare.py [-h] [-u] [--alpha UTEST_ALPHA]
{benchmarks,filters,benchmarksfiltered} ...
versatile benchmark output compare tool
<...>
optional arguments:
-h, --help show this help message and exit
-u, --utest Do a two-tailed Mann-Whitney U test with the null
hypothesis that it is equally likely that a randomly
selected value from one sample will be less than or
greater than a randomly selected value from a second
sample. WARNING: requires **LARGE** (9 or more)
number of repetitions to be meaningful!
--alpha UTEST_ALPHA significance level alpha. if the calculated p-value is
below this value, then the result is said to be
statistically significant and the null hypothesis is
rejected. (default: 0.0500)
```
Example output:
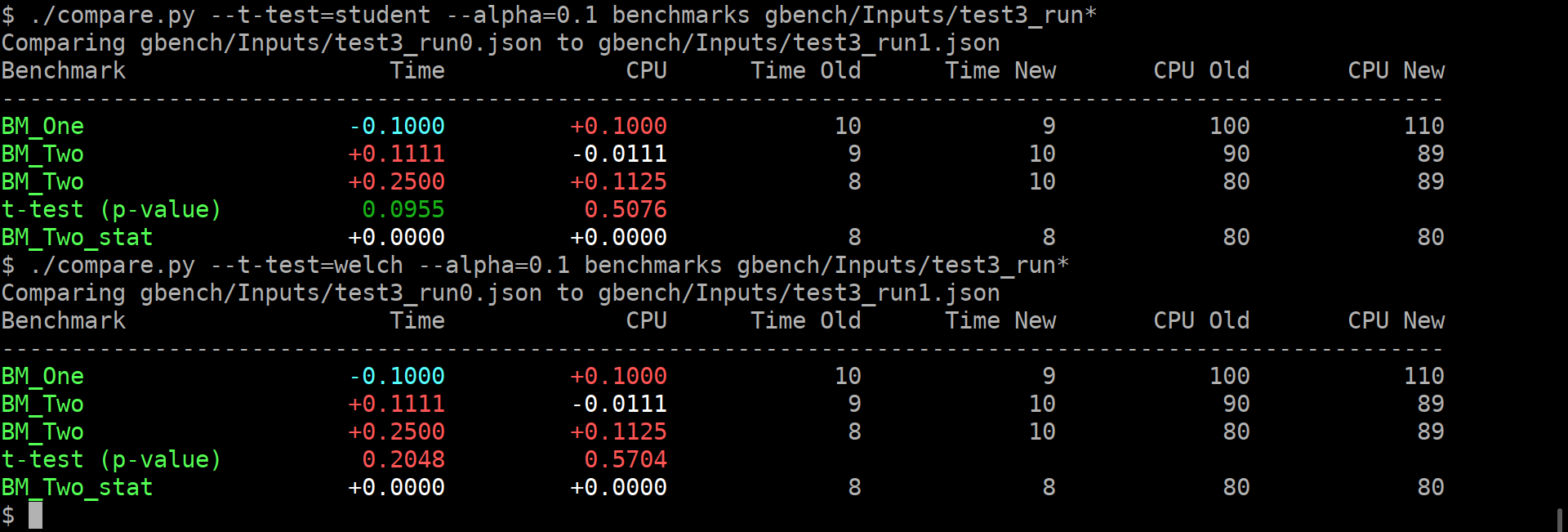
As you can guess, the alpha does affect anything but the coloring of the computed p-values.
If it is green, then the change in the average values is statistically-significant.
I'm detecting the repetitions by matching name. This way, no changes to the json are _needed_.
Caveats:
* This won't work if the json is not in the same order as outputted by the benchmark,
or if the parsing does not retain the ordering.
* This won't work if after the grouped repetitions there isn't at least one row with
different name (e.g. statistic). Since there isn't a knob to disable printing of statistics
(only the other way around), i'm not too worried about this.
* **The results will be wrong if the repetition count is different between the two benchmarks being compared.**
* Even though i have added (hopefully full) test coverage, the code of these python tools is staring
to look a bit jumbled.
* So far i have added this only to the `tools/compare.py`.
Should i add it to `tools/compare_bench.py` too?
Or should we deduplicate them (by removing the latter one)?
* Add tests to verify assembler output -- Fix DoNotOptimize.
For things like `DoNotOptimize`, `ClobberMemory`, and even `KeepRunning()`,
it is important exactly what assembly they generate. However, we currently
have no way to test this. Instead it must be manually validated every
time a change occurs -- including a change in compiler version.
This patch attempts to introduce a way to test the assembled output automatically.
It's mirrors how LLVM verifies compiler output, and it uses LLVM FileCheck to run
the tests in a similar way.
The tests function by generating the assembly for a test in CMake, and then
using FileCheck to verify the // CHECK lines in the source file are found
in the generated assembly.
Currently, the tests only run on 64-bit x86 systems under GCC and Clang,
and when FileCheck is found on the system.
Additionally, this patch tries to improve the code gen from DoNotOptimize.
This should probably be a separate change, but I needed something to test.
* Disable assembly tests on Bazel for now
* Link FIXME to github issue
* Fix Tests on OS X
* fix strip_asm.py to work on both Linux and OS X like targets
* [Tools] A new, more versatile benchmark output compare tool
Sometimes, there is more than one implementation of some functionality.
And the obvious use-case is to benchmark them, which is better?
Currently, there is no easy way to compare the benchmarking results
in that case:
The obvious solution is to have multiple binaries, each one
containing/running one implementation. And each binary must use
exactly the same benchmark family name, which is super bad,
because now the binary name should contain all the info about
benchmark family...
What if i tell you that is not the solution?
What if we could avoid producing one binary per benchmark family,
with the same family name used in each binary,
but instead could keep all the related families in one binary,
with their proper names, AND still be able to compare them?
There are three modes of operation:
1. Just compare two benchmarks, what `compare_bench.py` did:
```
$ ../tools/compare.py benchmarks ./a.out ./a.out
RUNNING: ./a.out --benchmark_out=/tmp/tmprBT5nW
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:16:44
------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------
BM_memcpy/8 36 ns 36 ns 19101577 211.669MB/s
BM_memcpy/64 76 ns 76 ns 9412571 800.199MB/s
BM_memcpy/512 84 ns 84 ns 8249070 5.64771GB/s
BM_memcpy/1024 116 ns 116 ns 6181763 8.19505GB/s
BM_memcpy/8192 643 ns 643 ns 1062855 11.8636GB/s
BM_copy/8 222 ns 222 ns 3137987 34.3772MB/s
BM_copy/64 1608 ns 1608 ns 432758 37.9501MB/s
BM_copy/512 12589 ns 12589 ns 54806 38.7867MB/s
BM_copy/1024 25169 ns 25169 ns 27713 38.8003MB/s
BM_copy/8192 201165 ns 201112 ns 3486 38.8466MB/s
RUNNING: ./a.out --benchmark_out=/tmp/tmpt1wwG_
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:16:53
------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------
BM_memcpy/8 36 ns 36 ns 19397903 211.255MB/s
BM_memcpy/64 73 ns 73 ns 9691174 839.635MB/s
BM_memcpy/512 85 ns 85 ns 8312329 5.60101GB/s
BM_memcpy/1024 118 ns 118 ns 6438774 8.11608GB/s
BM_memcpy/8192 656 ns 656 ns 1068644 11.6277GB/s
BM_copy/8 223 ns 223 ns 3146977 34.2338MB/s
BM_copy/64 1611 ns 1611 ns 435340 37.8751MB/s
BM_copy/512 12622 ns 12622 ns 54818 38.6844MB/s
BM_copy/1024 25257 ns 25239 ns 27779 38.6927MB/s
BM_copy/8192 205013 ns 205010 ns 3479 38.108MB/s
Comparing ./a.out to ./a.out
Benchmark Time CPU Time Old Time New CPU Old CPU New
------------------------------------------------------------------------------------------------------
BM_memcpy/8 +0.0020 +0.0020 36 36 36 36
BM_memcpy/64 -0.0468 -0.0470 76 73 76 73
BM_memcpy/512 +0.0081 +0.0083 84 85 84 85
BM_memcpy/1024 +0.0098 +0.0097 116 118 116 118
BM_memcpy/8192 +0.0200 +0.0203 643 656 643 656
BM_copy/8 +0.0046 +0.0042 222 223 222 223
BM_copy/64 +0.0020 +0.0020 1608 1611 1608 1611
BM_copy/512 +0.0027 +0.0026 12589 12622 12589 12622
BM_copy/1024 +0.0035 +0.0028 25169 25257 25169 25239
BM_copy/8192 +0.0191 +0.0194 201165 205013 201112 205010
```
2. Compare two different filters of one benchmark:
(for simplicity, the benchmark is executed twice)
```
$ ../tools/compare.py filters ./a.out BM_memcpy BM_copy
RUNNING: ./a.out --benchmark_filter=BM_memcpy --benchmark_out=/tmp/tmpBWKk0k
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:37:28
------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------
BM_memcpy/8 36 ns 36 ns 17891491 211.215MB/s
BM_memcpy/64 74 ns 74 ns 9400999 825.646MB/s
BM_memcpy/512 87 ns 87 ns 8027453 5.46126GB/s
BM_memcpy/1024 111 ns 111 ns 6116853 8.5648GB/s
BM_memcpy/8192 657 ns 656 ns 1064679 11.6247GB/s
RUNNING: ./a.out --benchmark_filter=BM_copy --benchmark_out=/tmp/tmpAvWcOM
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:37:33
----------------------------------------------------
Benchmark Time CPU Iterations
----------------------------------------------------
BM_copy/8 227 ns 227 ns 3038700 33.6264MB/s
BM_copy/64 1640 ns 1640 ns 426893 37.2154MB/s
BM_copy/512 12804 ns 12801 ns 55417 38.1444MB/s
BM_copy/1024 25409 ns 25407 ns 27516 38.4365MB/s
BM_copy/8192 202986 ns 202990 ns 3454 38.4871MB/s
Comparing BM_memcpy to BM_copy (from ./a.out)
Benchmark Time CPU Time Old Time New CPU Old CPU New
--------------------------------------------------------------------------------------------------------------------
[BM_memcpy vs. BM_copy]/8 +5.2829 +5.2812 36 227 36 227
[BM_memcpy vs. BM_copy]/64 +21.1719 +21.1856 74 1640 74 1640
[BM_memcpy vs. BM_copy]/512 +145.6487 +145.6097 87 12804 87 12801
[BM_memcpy vs. BM_copy]/1024 +227.1860 +227.1776 111 25409 111 25407
[BM_memcpy vs. BM_copy]/8192 +308.1664 +308.2898 657 202986 656 202990
```
3. Compare filter one from benchmark one to filter two from benchmark two:
(for simplicity, the benchmark is executed twice)
```
$ ../tools/compare.py benchmarksfiltered ./a.out BM_memcpy ./a.out BM_copy
RUNNING: ./a.out --benchmark_filter=BM_memcpy --benchmark_out=/tmp/tmp_FvbYg
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:38:27
------------------------------------------------------
Benchmark Time CPU Iterations
------------------------------------------------------
BM_memcpy/8 37 ns 37 ns 18953482 204.118MB/s
BM_memcpy/64 74 ns 74 ns 9206578 828.245MB/s
BM_memcpy/512 91 ns 91 ns 8086195 5.25476GB/s
BM_memcpy/1024 120 ns 120 ns 5804513 7.95662GB/s
BM_memcpy/8192 664 ns 664 ns 1028363 11.4948GB/s
RUNNING: ./a.out --benchmark_filter=BM_copy --benchmark_out=/tmp/tmpDfL5iE
Run on (8 X 4000 MHz CPU s)
2017-11-07 21:38:32
----------------------------------------------------
Benchmark Time CPU Iterations
----------------------------------------------------
BM_copy/8 230 ns 230 ns 2985909 33.1161MB/s
BM_copy/64 1654 ns 1653 ns 419408 36.9137MB/s
BM_copy/512 13122 ns 13120 ns 53403 37.2156MB/s
BM_copy/1024 26679 ns 26666 ns 26575 36.6218MB/s
BM_copy/8192 215068 ns 215053 ns 3221 36.3283MB/s
Comparing BM_memcpy (from ./a.out) to BM_copy (from ./a.out)
Benchmark Time CPU Time Old Time New CPU Old CPU New
--------------------------------------------------------------------------------------------------------------------
[BM_memcpy vs. BM_copy]/8 +5.1649 +5.1637 37 230 37 230
[BM_memcpy vs. BM_copy]/64 +21.4352 +21.4374 74 1654 74 1653
[BM_memcpy vs. BM_copy]/512 +143.6022 +143.5865 91 13122 91 13120
[BM_memcpy vs. BM_copy]/1024 +221.5903 +221.4790 120 26679 120 26666
[BM_memcpy vs. BM_copy]/8192 +322.9059 +323.0096 664 215068 664 215053
```
* [Docs] Document tools/compare.py
* [docs] Document how the change is calculated
* Tools: compare-bench.py: print change% with two decimal digits
Here is a comparison of before vs. after:
```diff
-Benchmark Time CPU Time Old Time New CPU Old CPU New
----------------------------------------------------------------------------------------------------------
-BM_SameTimes +0.00 +0.00 10 10 10 10
-BM_2xFaster -0.50 -0.50 50 25 50 25
-BM_2xSlower +1.00 +1.00 50 100 50 100
-BM_1PercentFaster -0.01 -0.01 100 99 100 99
-BM_1PercentSlower +0.01 +0.01 100 101 100 101
-BM_10PercentFaster -0.10 -0.10 100 90 100 90
-BM_10PercentSlower +0.10 +0.10 100 110 100 110
-BM_100xSlower +99.00 +99.00 100 10000 100 10000
-BM_100xFaster -0.99 -0.99 10000 100 10000 100
-BM_10PercentCPUToTime +0.10 -0.10 100 110 100 90
+Benchmark Time CPU Time Old Time New CPU Old CPU New
+-------------------------------------------------------------------------------------------------------------
+BM_SameTimes +0.0000 +0.0000 10 10 10 10
+BM_2xFaster -0.5000 -0.5000 50 25 50 25
+BM_2xSlower +1.0000 +1.0000 50 100 50 100
+BM_1PercentFaster -0.0100 -0.0100 100 99 100 99
+BM_1PercentSlower +0.0100 +0.0100 100 101 100 101
+BM_10PercentFaster -0.1000 -0.1000 100 90 100 90
+BM_10PercentSlower +0.1000 +0.1000 100 110 100 110
+BM_100xSlower +99.0000 +99.0000 100 10000 100 10000
+BM_100xFaster -0.9900 -0.9900 10000 100 10000 100
+BM_10PercentCPUToTime +0.1000 -0.1000 100 110 100 90
+BM_ThirdFaster -0.3333 -0.3333 100 67 100 67
```
So the first ("Time") column is exactly where it was, but with
two more decimal digits. The position of the '.' in the second
("CPU") column is shifted right by those two positions, and the
rest is unmodified, but simply shifted right by those 4 positions.
As for the reasoning, i guess it is more or less the same as
with #426. In some sad times, microbenchmarking is not applicable.
In those cases, the more precise the change report is, the better.
The current formatting prints not so much the percentages,
but the fraction i'd say. It is more useful for huge changes,
much more than 100%. That is not always the case, especially
if it is not a microbenchmark. Then, even though the change
may be good/bad, the change is small (<0.5% or so),
rounding happens, and it is no longer possible to tell.
I do acknowledge that this change does not fix that problem. Of
course, confidence intervals and such would be better, and they
would probably fix the problem. But i think this is good as-is
too, because now the you see 2 fractional percentage digits!1
The obvious downside is that the output is now even wider.
* Revisit tests, more closely documents the current behavior.
2373382284
reworked parsing, and introduced a regression
in handling of the optional options that
should be passed to both of the benchmarks.
Now, unless the *first* optional argument starts with
'-', it would just complain about that argument:
Unrecognized positional argument arguments: '['q']'
which is wrong. However if some dummy arg like '-q' was
passed first, it would then happily passthrough them all...
This commit fixes benchmark_options behavior, by
restoring original passthrough behavior for all
the optional positional arguments.
While the percentages are displayed for both of the columns,
the old/new values are only displayed for the second column,
for the CPU time. And the column is not even spelled out.
In cases where b->UseRealTime(); is used, this is at the
very least highly confusing. So why don't we just
display both the old/new for both the columns?
Fixes#425
* Json reporter: passthrough fp, don't cast it to int; adjust tooling
Json output format is generally meant for further processing
using some automated tools. Thus, it makes sense not to
intentionally limit the precision of the values contained
in the report.
As it can be seen, FormatKV() for doubles, used %.2f format,
which was meant to preserve at least some of the precision.
However, before that function is ever called, the doubles
were already cast to the integer via RoundDouble()...
This is also the case for console reporter, where it makes
sense because the screen space is limited, and this reporter,
however the CSV reporter does output some( decimal digits.
Thus i can only conclude that the loss of the precision
was not really considered, so i have decided to adjust the
code of the json reporter to output the full fp precision.
There can be several reasons why that is the right thing
to do, the bigger the time_unit used, the greater the
precision loss, so i'd say any sort of further processing
(like e.g. tools/compare_bench.py does) is best done
on the values with most precision.
Also, that cast skewed the data away from zero, which
i think may or may not result in false- positives/negatives
in the output of tools/compare_bench.py
* Json reporter: FormatKV(double): address review note
* tools/gbench/report.py: skip benchmarks with different time units
While it may be useful to teach it to operate on the
measurements with different time units, which is now
possible since floats are stored, and not the integers,
but for now at least doing such a sanity-checking
is better than providing misinformation.
This prevents errors when additional non-timing data are present in
the JSON that is loaded, for example when complexity data has been
computed (see #379).
This patch cleans up a number of issues with how compare_bench.py handled
the command line arguments.
* Use the 'argparse' python module instead of hand rolled parsing. This gives
better usage messages.
* Add diagnostics for certain --benchmark flags that cannot or should not
be used with compare_bench.py (eg --benchmark_out_format=csv).
* Don't override the user specified --benchmark_out flag if it's provided.
In future I would like the user to be able to capture both benchmark output
files, but this change is big enough for now.
This fixes issue #313.
* Change to using per-thread timers
* fix bad assertions
* fix copy paste error on windows
* Fix thread safety annotations
* Make null-log thread safe
* remove remaining globals
* use chrono for walltime since it is thread safe
* consolidate timer functions
* Add missing ctime include
* Rename to be consistent with Google style
* Format patch using clang-format
* cleanup -Wthread-safety configuration
* Don't trust _POSIX_FEATURE macros because OS X lies.
* Fix OS X thread timings
* attempt to fix mingw build
* Attempt to make mingw work again
* Revert old mingw workaround
* improve diagnostics
* Drastically improve OS X measurements
* Use average real time instead of max
This patch adds the compare_bench.py utility which can be used to compare the result of benchmarks.
The program is invoked like:
$ compare_bench.py <old-benchmark> <new-benchmark> [benchmark options]...
Where <old-benchmark> and <new-benchmark> either specify a benchmark executable file, or a JSON output file. The type of the input file is automatically detected. If a benchmark executable is specified then the benchmark is run to obtain the results. Otherwise the results are simply loaded from the output file.